Gene Editing with Python
On this episode, you'll meet David Born, a computational biologist who uses Python to help automate genetics research and helps move that work to production.
Episode Deep Dive
Guest Introduction and Background
David Born is a computational biologist at Beam Therapeutics, where he works on using Python and large-scale computing to advance gene editing research. He started from the biology side of the world, earned his graduate degree studying genetics, and then picked up Python to tackle complex data analysis tasks. At Beam Therapeutics, David focuses on developing software pipelines and infrastructure to automate and scale genetics research, including CRISPR-based gene editing data. His day-to-day involves analyzing vast sequencing datasets, collaborating with lab researchers, and orchestrating large workloads in the cloud.
What to Know If You're New to Python
This episode touches on how Python is leveraged in biology and big data contexts. Having a basic grasp of running scripts, working in notebooks, and understanding packages like NumPy and pandas can be helpful. Many of David’s workflows rely on standard Python practices—such as version control, containerization with Docker, and an understanding of web frameworks like Django—to ensure reproducibility and scalability. Here are a resource to deepen your knowledge:
- Documentation for pandas and NumPy: Focus on data manipulation and fundamental numeric operations.
Key Points and Takeaways
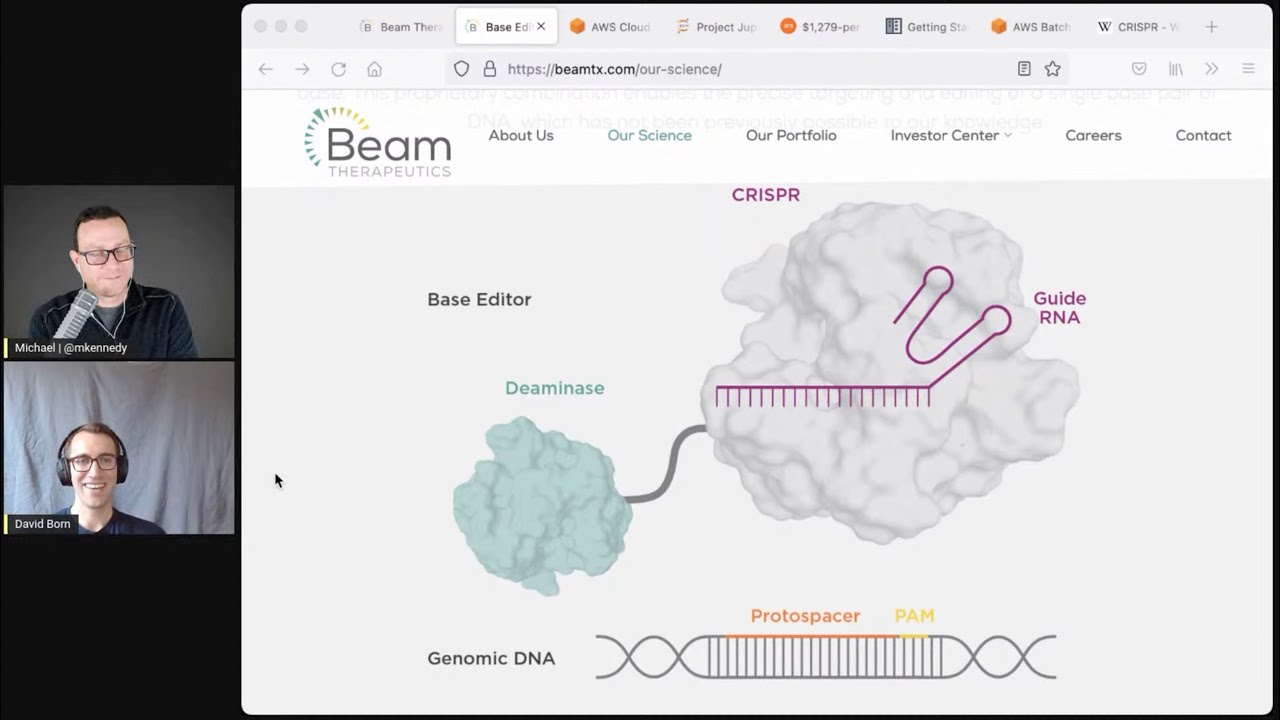
- Precision Gene Editing and CRISPR CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) is a molecular machine that can be directed to precisely edit specific DNA sequences. David discusses how beam therapies target single-gene mutations—such as those involved in sickle cell disease—and use CRISPR-based approaches to correct them. The complexity of tracking, validating, and scaling these gene edits leads to massive data demands, making Python’s tooling invaluable.
- Tools / Links:
- Managing Big Data in Genetics Gene editing experiments produce large sequencing datasets that must be processed for accuracy, reproducibility, and compliance. Python’s data science ecosystem (e.g., pandas, NumPy, Jupyter notebooks) lets researchers quickly prototype analysis steps. Once validated, these steps become more formalized pipelines for production use.
- Tools / Links:
- AWS Cloud Infrastructure for Scalability To handle massive sequencing runs—sometimes tens of gigabytes to terabytes of data—Beam Therapeutics uses AWS services extensively. David highlighted using S3 for data storage, AWS Lambda for event-driven tasks, and AWS Batch for highly parallel workloads. This serverless and on-demand model drastically simplifies managing thousands of compute cores.
- Tools / Links:
- Infrastructure as Code with AWS CDK One of the pivotal components enabling reproducible and maintainable cloud infrastructure is the AWS Cloud Development Kit (CDK) in Python. Rather than hand-configuring services through the AWS console, David’s team stores infrastructure definitions in source control and programmatically deploys them.
- Tools / Links:
- Orchestrating Pipelines with Luigi and Other Workflow Managers Large bioinformatics pipelines often involve chaining together many third-party tools and custom Python scripts. David’s team leans on Luigi for Python-based workflows. Other popular options mentioned are Nextflow, Airflow, and DAXter, each providing a system to build complex directed acyclic graphs (DAGs) of tasks.
- Tools / Links:
- Reproducibility in Computational Biology Biotechnology and pharmaceutical research must be reproducible for regulatory and scientific reasons. David highlighted how Docker containers, pinned library versions, and source-controlled data workflows help ensure consistent results. They also build pipelines so that external partners—such as the FDA or other collaborators—can rerun analyses with the same code.
- Tools / Links:
- docker.com
- pypi.org for version pinning
- Tools / Links:
- From Jupyter Prototypes to Production Many data experiments begin as quick prototypes or proof-of-concepts in Jupyter notebooks. Once validated, the code moves into more robust scripts with standardized libraries, eventually placed into orchestrated Docker containers for large-scale runs. This systematic process ensures a smooth transition from data exploration to production pipelines.
- Tools / Links:
- Django and Database Choices for Genetic Data David’s team uses Django with a MySQL backend to manage large amounts of metadata and experimental records. While some worry about ORMs slowing down, their well-indexed schema and optimized queries have kept performance high. The Django REST Framework also helps expose internal services as needed.
- Tools / Links:
- Scaling Out HPC Jobs Occasionally, the team needs truly massive CPU resources—10,000+ cores for days—to simulate or analyze certain molecular events. Rather than buying a supercomputer, AWS Batch orchestrates ephemeral EC2 instances in a cluster of Docker containers. They carefully test pipeline logic on smaller subsets of data to avoid expensive mistakes on large-scale runs.
- Tools / Links:
- Remote Development with VS Code On a day-to-day basis, many of the scientists use Visual Studio Code’s remote development extension to interact directly with cloud VMs or on-prem machines. This setup allows them to maintain a consistent Python environment, run computations on more powerful servers, and keep iteration loops tight.
- Tools / Links:
- Team Collaboration and Cost Management Large bioinformatics computations can cost tens of thousands of dollars. David emphasized the importance of collaboration between experimental scientists and software engineers, plus frequent checks and tests for correctness before launching large runs. With good communication and versioning, they keep mistakes and wasted compute to a minimum.
- Tools / Links:
Interesting Quotes and Stories
- Big-Picture Vision: David mentioned, “One day we could say, ‘Hey Alexa, how do we cure sickle cell disease?’ and it might recommend the gene editing steps to fix it.” This underscores the future-facing approach at Beam Therapeutics.
- High-Performance Computing Scare: There was a story about how a code misconfiguration can bring down entire clusters or fill up a disk quickly. It reminds everyone to test carefully and version-check your pipelines before a large-scale run.
- Crossing Biology and Software: David described how coming from a purely biological background, it took just a few weeks to become productive in Python—reinforcing how accessible it can be for domain experts in any field.
Key Definitions and Terms
- CRISPR: A molecular tool allowing precise DNA sequence targeting and cutting for gene editing.
- AWS Batch: A managed service to run hundreds or thousands of containerized batch computing jobs at scale on AWS.
- Luigi: A Python-based workflow manager for building complex data processing tasks (DAGs).
- ORM (Object Relational Mapper): A layer allowing you to interact with databases via class-based APIs, such as Django’s models, rather than raw SQL.
- Docker Container: A lightweight virtualization method that bundles code, runtimes, and dependencies into portable units.
Learning Resources
- Python for Absolute Beginners: Get up to speed quickly if you’re new to Python or programming in general.
- Data Science Jumpstart with 10 Projects: Build a strong foundation for data-driven tasks, which is especially relevant to large-scale bioinformatics.
- Django: Getting Started: Learn how to build robust backends for data management, perfect if you want to replicate Beam’s Django + MySQL approach.
- Fundamentals of Dask: While David’s team used AWS Batch, Dask is another valuable tool for scaling Python workflows and computations.
Overall Takeaway
Python has become central to the future of gene editing research, as seen in Beam Therapeutics’ efforts to tackle enormous genetics datasets with robust and reproducible pipelines. Through a combination of established Python libraries, containerization, and cloud-based scale-out infrastructure, teams like David Born’s can quickly iterate on new genetic insights and move proven methods into production. This episode highlights not only the cutting-edge science but also the power of Python to bring domain experts and software engineers together to solve real-world problems—potentially changing healthcare and saving lives in the process.
Links from the show
Beam Therapeutics: beamtx.com
AWS Cloud Development Kit: aws.amazon.com/cdk
Jupyter: jupyter.org
$1,279-per-hour, 30,000-core cluster built on Amazon EC2 cloud: arstechnica.com
Luigi data pipelines: luigi.readthedocs.io
AWS Batch: aws.amazon.com/batch
What is CRISPR?: wikipedia.org
SUMMIT supercomputer: olcf.ornl.gov/summit
Watch this episode on YouTube: youtube.com
Episode #335 deep-dive: talkpython.fm/335
Episode transcripts: talkpython.fm
---== Don't be a stranger ==---
YouTube: youtube.com/@talkpython
Bluesky: @talkpython.fm
Mastodon: @talkpython@fosstodon.org
X.com: @talkpython
Michael on Bluesky: @mkennedy.codes
Michael on Mastodon: @mkennedy@fosstodon.org
Michael on X.com: @mkennedy
Episode Transcript
Collapse transcript
00:00 Gene therapy holds the promise to permanently cure diseases that have been considered
00:04 lifelong challenges, but the complexity of rewriting DNA is truly huge and lives in its
00:09 own special kind of big data world. On this episode, you'll meet David Born, a computational
00:15 biologist who uses Python to help automate genetics research and helps move that work
00:20 to production. This is Talk Python To Me, episode 335, recorded September 15th, 2021.
00:27 Welcome to Talk Python To Me, a weekly podcast on Python. This is your host, Michael Kennedy.
00:44 Follow me on Twitter where I'm @mkennedy and keep up with the show and listen to past episodes
00:49 at talkpython.fm and follow the show on Twitter via at talkpython. We've started streaming most
00:56 of our episodes live on YouTube. Subscribe to our YouTube channel over at talkpython.fm
01:00 slash YouTube to get notified about upcoming shows and be part of that episode. This episode
01:07 is brought to you by Shortcut, formerly known as clubhouse.io, and us over at Talk Python
01:12 Training. And the transcripts are brought to you by Assembly AI. David, welcome to Talk Python
01:18 to Me.
01:18 Thanks, Michael. It's great to be here.
01:19 Yeah, it's great to have you here. One of the things I really love to explore is the
01:24 somewhat non-traditional use cases of Python that are not straight down the, I'm building
01:30 an API and something that talks to a database or a startup, right? Something like that, but
01:35 blending it with other technologies and science and whatnot and genetics plus Python, it's going
01:41 to be interesting.
01:42 It sure is.
01:43 When you got into this, did you start out on the biology side or the programming side of
01:48 the world?
01:48 I definitely started out on the biology side. Yeah. So I went all the way through grad school
01:53 with relatively minimal formal programming experience.
01:57 Yeah. Okay. So you studied biology and genetics and whatnot. And then how'd you end up here
02:03 on a Python podcast?
02:04 Yeah. So I always thought that programming would be cool, but I didn't really have much of an
02:09 opportunity through my undergraduate studies to really do much formal programming. I took our one
02:15 computer science class that my college had to offer. It was in C++. I think I wrote a Boggle program,
02:21 something with some recursion in there. It was pretty fun. Didn't really get to use Python until
02:26 graduate school. I was in a genetics course and we were basically tasked with doing some data analysis
02:33 on published data and then reproducing some plots in a figure, then extending it further.
02:38 My partner and I decided to learn Python, teach it to ourselves so that we could do this. We heard that
02:45 it was a good way to do data analysis in biology. And so we basically taught it to ourselves and we used
02:52 NumPy, some basic string searching and things to redo this analysis. And it was really amazing what we could
03:00 do with Python for that civil project.
03:02 That's awesome. How'd the learning project go? Like coming from not having a ton of programming,
03:06 what was your experience like?
03:08 It was relatively easy. I would say I think my brain sort of fits pretty well with how
03:13 programming languages work, but it was definitely a lot in a short amount of time to really dive into
03:18 how to make sure your while loops don't stay open. And then someone tells you maybe you shouldn't use
03:24 a while loop at all. I was learning a lot of things not to do right away.
03:28 Yeah, of course. But you had to get all the analysis done, right? So you got to solve all the problems
03:33 and power through.
03:34 Yeah, we did. We got our analysis done there and we got some, we reproduced some plots and got some
03:40 new analysis made. And I think we really just, I really came to appreciation of how much you can do
03:45 in a short amount of time with just a little bit of coding knowledge or essentially none.
03:49 Yeah. I think that's really an important takeaway that a lot of people, you know, maybe many people
03:54 listen to podcasts already kind of know, but I think looking in from the outside, it feels like,
03:59 oh, I've got to, you know, go get a degree in this to be productive or useful. And really what you need
04:06 is like a couple of weeks and a small problem and you're already already there.
04:10 Absolutely. Yeah. I've all, I've definitely found that just learning through, through doing has been
04:15 the way I've, I've worked entirely. I have essentially no formal programming training, no,
04:20 no coursework and I'm using Python every day.
04:23 Yeah. That's fantastic. Yeah. I didn't take that much computer science and college either. Just
04:27 enough to do the extra stuff from my math degree. Very cool. All right. Now how about today? You're
04:32 working at Beam Therapeutics doing genetic stuff. Tell us about what you do day to day.
04:37 Yeah. So I'm on the computational sciences team at Beam Therapeutics. We're a gene editing company. So
04:44 we develop these precision genetic medicines that are, we're trying to develop them to cure genetic
04:50 diseases that are caused by single genetic changes in the genome.
04:56 Like a point shift mutation or something like that.
04:59 Yep. Yep. And so if you have one of these genetic changes, you might have a disease that is lifelong
05:06 and there aren't any cures for most of these diseases. So we're trying to create these, we call
05:12 them hopefully lifelong cures for patients by changing the genetic code back to what it should be.
05:18 That's incredible. It seems really out of the future. I mean, it's, I think it's one thing to
05:24 understand genetics at play and it's even amazing to be able to read the gene sequences, but it's
05:31 entirely another thing I think to say, and let's rewrite that.
05:34 Yeah. We're definitely at the cutting edge of a lot of biotechnology and science that has really come
05:41 to have had in the last decade with the, with CRISPR and technologies that use CRISPR, which ours do be able to precisely target genetic sequences. It's a really fascinating place to work and it's a privilege.
05:54 I bet a lot of people go to work and they end up writing what you might classify as forms over data. It's like, well, I need a view into this bit of our database or I need to make, be able to run a query to just see who's got the most sales this week or, you know, something like that.
06:10 And that's important work and it's, it's useful and there's cool design patterns and whatnot you can focus on, but it's also, it's not like what people dream of necessarily building when they wake up.
06:21 But this kind of science, like maybe so, right? Like these are the really interesting problems that both have a positive outcome, right? You're helping cure disease, not just, you know, shave another one, one hundredth of a percent off of a transaction that you get to keep or, you know, something like that. Right. From like in finance.
06:39 Yeah.
06:39 You get to use really cool tech to do it too, like programming wise.
06:42 Yeah. I mean, one of our dreams that we joke about on the computational team is that like it's conceivable one day we could, you know, say like, Hey Alexa, how do we cure sickle cell disease? And it'll tell you what parts of our technology we should put together to cure that disease. That's sort of like the pipe dream of, of where we could go if we combine all our data in the right ways. And I think all of the stuff we'll talk about today is really just laying out the framework for that.
07:09 Yeah, absolutely. So you mentioned CRISPR. Maybe tell people a bit about that biotechnology.
07:14 Yeah. So CRISPR is molecular machine, which a lot, which targets a very specific place in a specific genetic sequence. And so usually people are using CRISPR to target a specific place in the genome, a specific sequence. And what CRISPR does naturally is to cut at that sequence.
07:38 So it'll cut in a very specific place in the genome. And as people were using CRISPR, we could actually decide where it's going to cut by giving it a different targeting sequence. This sort of like directed molecular machine is a basis of a whole new field of biotechnology using CRISPR and CRISPR derived technology.
08:00 So our technology is like kind of a CRISPR 2.0 where we don't use CRISPR to cut. We use the localization machinery and we add onto it another protein, which just changes a base instead of cutting the DNA itself.
08:16 So it's slight variation, but it's still using this same CRISPR technology.
08:20 Okay. So let me see if my not very knowledge filled background understand here is kind of a, has a decent analogy. So does it work basically like you give it like almost like find and replace, like you give it a sequence of pairs and it says, okay, if I find a TTCAT, like enough specificity and that it's like, that's the unique one. And then it does like cut at that point. Is that kind of how it works?
08:46 Yep. It's pretty much just like that. We give it the sequence you want to target. And then if it finds that sequence in the genome, it will cut the genetic material, the DNA at that position for normal CRISPR.
09:00 When you're coming up with these and you say, we're going to rewrite the DNA to solve this problem. How do you do it on a large enough scale? Right. Like how do you, I mean, how much of the body has to be changed for this to be kind of, to be permanent, right?
09:14 Right. It's definitely a tricky question. I think we are, we definitely leverage human biology, how the human body works for a lot of these problems. For example, our, some of our leading drug candidates are for sickle cell disease. And because of the way sickle cell disease manifests in red blood cells and red blood cells are created through a set of, through, there's a specific type of cell that creates red blood cells.
09:41 And we can, if you access that type of cell and you cure, cure a sickle cell in these progenitor cells, the stem cells, then you can create all red blood cells from a cured population.
09:54 So if you can target it to the regenerative cells, you can cure sickle cell throughout the body essentially, because the symptoms are from red blood cells. There's a lot of diseases that, which by curing a single organ, you can cure the symptoms of the disease because that's where it actually manifests.
10:13 Right. Like, like, like diabetes or sickle cell anemia or something like that?
10:17 Yeah. Like sickle cell diseases in the liver, like some blindnesses in the eye. By just targeting specifically where the symptoms occur, you can cure the disease.
10:27 Yeah. That's amazing. Is it a shot? Is it a blood transfusion or like, how do you deliver this stuff?
10:32 Yeah. Delivery is a huge place of research and it definitely depends on the type of targeting we're doing. So for something in the eye, it would probably be an injection.
10:41 For something in the blood, it's slightly more complicated and it's not an injection. For something in the liver, it would probably be more akin to a injection or dosing regimen.
10:54 Okay. It sounds really fascinating. Like I said, it feels like it's a little bit out of the future to be able to come up with these and just say, no, we're just going to rewrite this little bit of the genetics and I think it'll be good. But if you can make it happen, it's pretty clear how it's obviously a benefit, right? Like those progenitor cells, they eventually have to recreate new ones. And then the way they do that is they clone their, their current copy of the DNA, which is the fixed one, right? So if you can get enough of them going, it'll just sort of propagate from there.
11:22 Yep. And we're also the benefit that sometimes you only need to cure a small fraction to remove the symptom. So lots of things going for us.
11:31 That's awesome. So I'm sure there's a lot of people involved in this type of work. What exactly are you and your team working on?
11:39 Yeah. So our team, we call ourselves the computational sciences team. We really sit in the, in the middle of the research and development arm of the organization, processing all of our sequencing data and some other data as well. And as you can imagine, with our technology changing DNA, changing genomes, there's a lot of sequencing data because what we're trying to do is change a genetic sequence.
12:07 So we have to read out that genetic sequence and then figure out has it changed, how many copies are changed and things like that.
12:14 Yeah.
12:14 Yeah.
12:14 The field of the techniques of next generation sequencing NGS are pretty broad. And we deal with a lot of different types of these next generation sequencing assays that are being done at the end. Our team really processes, analyzes, and collaborates with the experimental scientists on performing and developing these experiments.
12:36 Cool. So the scientists will do some work and they'll attempt to use CRISPR-like technology to make changes. And then they measure the changes that they've made. And it's, you all sort of take that data and compare it and work with it.
12:50 Right. Yeah. The act of measuring the changes itself is relatively computationally intensive. So we run and support pipelines for all of these standard assays as well, which is, is part of our, our job.
13:03 How much data is in DNA? I mean, I know that human biology stores or just biology stores an insane amount of data, but also computers store an insane amount of data and processing. So I'm not sure like which, where the sort of trade-off is, but what sort of data are we talking about? How much?
13:20 Yeah. I mean, it definitely depends on the type of assay that we're doing, like the scale of the data. I would say usually we're not looking, sometimes we're looking at things that are looking at all the bases in the genome. More often we're looking at defined regions of the genome where we're trying to make the change.
13:39 Right. You're like, this one gene is the problem, right? And let's look at that.
13:42 Right. Yeah. We're trying to target here. So that's where we're looking. It definitely depends on the assay, but, but I guess in terms of data scale, in terms of file sizes, perhaps that would be accessible.
13:52 Yeah.
13:54 For standard things, it would be on the order of a few gigabytes per experimental run. For some of our larger assays, it's 10 to 100 times that.
14:04 Right.
14:04 Per experiment.
14:05 That's a lot of data, but not impossible to transmit sort of amounts of data, right? Or store.
14:10 Right. Every little piece of it is, is pretty manageable. When you start combining them together and looking at some doubt, like your downstream results of things, the data does get pretty, pretty large.
14:22 But I wouldn't say we're at the scale of like big data analytics at Google or anything like that.
14:26 Yeah. Yeah. Or the LHC. If you've ever looked at the data flow layers at LHC, it's like the stuff near the collectors is just unimaginable amounts of data. Right.
14:36 Yeah. I haven't looked, but I'm sure it's certainly.
14:39 Yeah. They've got a lot of stuff built in, like onboard the detectors to filter it down a bunch. And then it goes for processing. Then it gets filtered down some more. And then like eventually it gets to a point where like now we have enough space.
14:51 on hard drives to save it.
14:53 But before that, it was like, it couldn't be saved on hard drives.
14:56 It was just too much.
14:56 Yeah, very interesting.
14:58 But it sounds to me like some of the real big challenges for you all is the computational bits, right?
15:03 Because if you take all these things and you can end up in like combinatorial comparison scenarios,
15:09 I'm sure that that can really blow up the computing time.
15:12 Yeah, I think definitely there's a lot of the challenges sometimes in these combinatorics.
15:18 And there's also just a lot of, there's steps that have to go on to process a lot of this data.
15:24 There's a lot of biology specific pieces of software that we use for various things.
15:30 And we have to string them together to create a complete data processing pipeline.
15:37 And a lot of that art is part of our job, how to get all these tools to behave together
15:42 and act in unison to actually process the data effectively.
15:46 I can just imagine some of these pipelines are tricky.
15:49 It's like, well, okay, so it starts over here where the robot gets it and it reads this data
15:54 and we can access that off the hard drive.
15:56 And then it has to be sent to this Windows app that actually doesn't have an API.
16:00 So we got to somehow like automate that thing and then get data out of it.
16:05 And then is that what it's like?
16:06 That's some of it for sure.
16:07 Tell us about your life.
16:08 Okay.
16:09 Yeah.
16:09 I mean, there's a couple aspects and I think we might touch on them, but for sequencing experiments,
16:14 the pipelines are more defined because we usually get the data from a source that's already in the cloud,
16:19 which I'm always happy about.
16:21 If we can start in the cloud, we'll stay in the cloud and that's a nice place to be.
16:24 For data that's coming directly from instruments on premises, there is another layer of art
16:32 that has to do with software on the instrument, software that gets the data to the cloud
16:37 and moves it around between our other database sources.
16:41 And that is a fun project in itself there.
16:43 Yeah.
16:44 I can imagine.
16:44 I've worked in some areas where it's like collecting data from all these different things
16:49 and then, you know, to process it and move it along.
16:52 And yeah, it's not always just send it from this Python function to that Python function
16:57 to that Python function.
16:58 There's a lot of janky stuff that wasn't really meant to be part of pipelines possibly.
17:03 Absolutely.
17:04 Yeah.
17:06 This portion of Talk Python To Me is brought to you by Shortcut, formerly known as clubhouse.io.
17:11 Happy with your project management tool?
17:13 Most tools are either too simple for a growing engineering team to manage everything
17:18 or way too complex for anyone to want to use them without constant prodding.
17:22 Shortcut is different though, because it's worse.
17:25 No, wait, no, I mean it's better.
17:26 Shortcut is project management built specifically for software teams.
17:30 It's fast, intuitive, flexible, powerful, and many other nice positive adjectives.
17:35 Key features include team-based workflows.
17:37 Individual teams can use default workflows or customize them to match the way they work.
17:42 Org-wide goals and roadmaps.
17:44 The work in these workflows is automatically tied into larger company goals.
17:48 It takes one click to move from a roadmap to a team's work to individual updates and back.
17:54 Type version control integration.
17:56 Whether you use GitHub, GitLab, or Bitbucket, clubhouse ties directly into them
18:00 so you can update progress from the command line.
18:03 Keyboard-friendly interface.
18:04 The rest of Shortcut is just as friendly as their power bar, allowing you to do
18:09 virtually anything without touching your mouse.
18:11 Throw that thing in the trash.
18:13 Iteration planning.
18:14 Set weekly priorities and let Shortcut run the schedule for you with accompanying burndown charts
18:20 and other reporting.
18:20 Give it a try over at talkpython.fm/shortcut.
18:25 Again, that's talkpython.fm/shortcut.
18:29 Choose shortcut because you shouldn't have to project manage your project management.
18:33 with robots, do you have to actually talk to the robots and like any of the
18:41 type of automated things?
18:42 Yeah, for lab automation is what we call our team that has the robots as we like to
18:49 call them.
18:50 As you can imagine with a lot of these types of experiments, they can be
18:54 made much more efficient if we can have robots doing the actual transfer of liquids
18:59 and incubation and centrifugation, these scientific techniques that sometimes you need
19:06 someone in the lab to do, but oftentimes you can automate them.
19:09 So the lab robotics aspect is an important part of how we can efficiently generate
19:15 data.
19:16 A lot of the issues around that come with how to pass instructions to the instrument
19:22 and how to get back data from the instrument when it's done.
19:26 and then there's a whole other art of making the instruments actually orchestrate
19:30 together, which is held in a different world of software.
19:34 I don't actually work on that part myself.
19:36 Yeah, that part of programming definitely seems a little bit magical, getting factories
19:41 or automations of different machines to work together.
19:44 It's very cool.
19:45 There's a whole lot of proprietary software involved in actually running
19:49 the instruments, but in terms of the problems in getting the data to them,
19:53 I think it's one of those, it could be a relatively common software problem
19:57 of getting information from somewhere in the cloud.
20:01 We have electronic lab notebooks and laboratory data systems that are in the
20:07 cloud and users will be submitting information about how they want their samples
20:12 processed by the robots.
20:14 There's the problem of getting that to the instruments themselves.
20:18 And I think it actually sort of reminded me of the talks you guys had in
20:22 episode, I think it was 327, the small automation projects.
20:26 We end up with quite a few of those here where we have these relatively small
20:31 tasks of take data from an API, put it somewhere where a robot can access it.
20:37 Usually we use AWS S3, and these sort of very small data handling tasks end up
20:44 being these nice little projects for Python to come into play.
20:47 Awesome.
20:48 I can see that that definitely happens, right?
20:51 if a file shows up here, grab it, upload it to that bucket, and name it whatever the
20:56 active experiment is with the data or something like that, right?
20:59 That's a very small program to write, but then that becomes a building block
21:03 in this flow, putting these pieces and machines together.
21:06 Right, yeah.
21:07 Once it comes into the cloud, we do another set of data processing on it,
21:12 we upload it to our current databases, and all of that we orchestrate on
21:18 AWS using, I guess they call it their serverless design patterns, so we don't have to
21:24 handle anything on our own computers.
21:26 Yeah, that's really nice, and the serverless stuff probably helps you avoid
21:30 running just tons of VMs in the cloud, right?
21:33 Like, it can all be on demand, like the Lambda trigger is a file appears in this
21:37 S3 bucket, so then it starts down the flow, right?
21:40 Absolutely, yeah.
21:41 I really don't like maintaining a lot of infrastructure, although we do have,
21:44 a good amount of it that we do have to maintain, I find that these small Python
21:49 functions are the perfect use case for those event-driven Lambda functions,
21:55 which are running these very simple pieces of code.
21:58 When an object appears in S3, they get a small event about when the object
22:03 was uploaded, and then they do their thing, a little bit of data conversion,
22:07 send it to an API, and now the data is in our data store.
22:11 And those things just happen, and they're super consistent, they don't require anything
22:16 on my end to maintain.
22:18 It's a pretty beautiful pattern.
22:20 That's awesome.
22:20 They don't need things like, oh, there's a new kernel for Linux that patches a
22:26 vulnerability, so let's go and patch our functions, right?
22:30 Just it's all magic.
22:31 It all happens on its own.
22:32 Right.
22:32 It does feel like magic a lot of the time, yeah.
22:35 Setting them up can be a little bit of a challenge, but once they're there, they are
22:39 very consistent.
22:40 Yeah, yeah, cool.
22:40 One of the things you talked about doing, you seen, was the AWS CDK, or Cloud
22:46 Development Kit.
22:47 I think I've heard of this before, but it's like, I've definitely not used this
22:51 personally.
22:51 Tell us, what is this?
22:53 How does it help you?
22:53 Right, so when I started dabbling with these Lambda functions, what I found pretty
23:00 soon after I had four or five of these functions, which I had uploaded on the
23:06 AWS Council, and if anyone's used that, know that it can be pretty tedious once
23:11 you have a few things running, getting all your permissions and things set up.
23:15 And I was looking for better ways to do this, and at that point, it was early
23:21 days for this AWS Cloud Development, AWS CDK, and what it is, is a way to write
23:28 your infrastructure as code in essentially any common language, programming language.
23:35 write JavaScript, TypeScript, I think you can do Java, Go, and Python perhaps.
23:40 And using it, you can define your cloud infrastructure in an object-oriented
23:47 way with all the parameters they need, and you can deploy it to different AWS
23:52 accounts, you can take it down, you can reconfigure it, you can look at how it's changed
23:57 since your last commit, and you can store all that configuration in source
24:01 control.
24:02 And this has allowed us to scale up, in terms of number of Lambda functions,
24:07 I think we have maybe 60 or something now, which would be unmaintainable
24:12 in the console, but they're essentially just completely maintained inside of
24:16 source control these days using AWS CDK and Python.
24:20 Yeah, it seems super neat.
24:21 I have not used it, but on the page, which I'll link to in the show notes,
24:25 they have Werner Vogels, the CTO of Amazon, AWS, and talks about some of the benefits and
24:34 kind of how it all fits together.
24:35 And like you said, you can store your cloud structure definition in source control.
24:40 You can run unit tests against your infrastructure to say, if I apply all these commands
24:44 to AWS, so I actually get what I was hoping to get out of it.
24:48 yeah, it seems like a really neat thing for this infrastructure's code bits.
24:53 Yeah, and I think it definitely really shines when you're developing larger pieces of
24:58 infrastructure, but I would encourage people to check it out, even if they
25:02 have a small automation type project.
25:05 This is what I was thinking of when I was listening to episode 327 the other day.
25:10 When you have these things you want to run on your computer with a cron job, you
25:15 can actually run them for free on AWS Lambda.
25:19 You get a bunch of free time on the free tier, and you try it out.
25:23 You don't need to make sure your system D processor for whatever is running, and it's
25:29 a pretty cool way to get familiar with how to do some of these things on AWS.
25:33 I'm not sure if this also exists for other cloud providers.
25:37 We use AWS in particular, so that's what I know, but it might also exist
25:41 for things like Azure and Google Cloud.
25:43 Sure.
25:44 Yeah, maybe.
25:44 I don't know if it does either, but it definitely seems useful for what you all are doing
25:48 there.
25:48 One of the things it could be useful for, and one of the challenges I suspect
25:52 that you run into more than a lot of places, certainly more than like an e-commerce
25:56 site or something like that, is reproducibility.
25:59 If you're going to say, we're going to come up with a treatment that literally gets
26:04 injected into people, you've got to go through FDA, you've got to have possibly peer-reviewed
26:11 stuff.
26:11 There's a lot of reproducibility and data stewardship going on, I imagine, right?
26:17 Absolutely.
26:18 That's definitely the case.
26:19 We are very cognizant of the fact that we have to make our software as reproducible as
26:25 possible.
26:25 I think a lot of that lends itself to just using good development practices,
26:30 containerizing everything that you're doing for data processing, getting all your
26:35 versions, appropriate source control, all of these things.
26:40 And the infrastructure is definitely a piece of that because if we can't deploy the
26:45 pipeline in the same way a year from now, we won't be able to get the same
26:50 results from the data.
26:51 And that's a problem.
26:53 That's a big challenge.
26:54 It's tricky, right?
26:55 Because things like containers and source control, they'll absolutely get you
27:00 very far.
27:00 But then you're also depending on these external things that have been very
27:06 stable and very likely will be, like Lambda, for example, but what if Amazon went out
27:11 of business, right?
27:12 Which, you know, is kind of laughable, right?
27:15 It's only like doubled its revenue this year or something.
27:17 But, you know, theoretically, it's possible that AWS decided to shut down
27:22 or something like that, right?
27:24 Yeah, it's definitely true.
27:25 So you got to think, these are trade-offs, right?
27:27 But at the same time, it's so enabling for you to just scale out all this workload.
27:31 Yeah, I mean, in terms of how we create data pipelines, I think a lot of
27:36 the ways we do it, we do have to be aware of creating them in a way that
27:40 you can run them outside of the cloud.
27:43 Sometimes we need to allow a third party to run our data analysis in a regulated way.
27:49 And that requires us to have essentially running it internally, we run it in the
27:55 cloud, which is efficient for scaling, but we also need to be able to take that
27:59 same piece of software and run it in a way that may not be on AWS, may not be in the
28:04 cloud at all.
28:05 And that creates some interesting software challenges.
28:08 Yeah, I'm sure, because so many of the APIs are, you know, cloud native,
28:12 right?
28:12 You import Bodo, Bodo do this thing, or something like that, right?
28:17 How do you deal with that?
28:18 I think a lot of it comes down to structuring your workflows and your data pipelines in
28:25 ways that are, they're not really using the cloud as much as they're, they're using the
28:29 cloud as a, well, the file system and a way to gain compute, but they're not
28:35 using any piece of the cloud to actually affect the data processing itself.
28:39 So ideally, if you create the data pipeline in a way that is appropriate,
28:44 you can both run it in the cloud and run it locally and achieve identical
28:49 results because it's containerized.
28:52 Primarily, that's the reason, I guess.
28:54 Yeah.
28:54 But doing that, I think, is a challenge in itself.
28:57 It's really like the process of getting a data pipeline built is a big ask in our
29:03 field.
29:03 I think a lot of companies spend a lot of time thinking about this.
29:06 Yeah.
29:07 I'm sure they do.
29:08 Getting it right is a huge enabler, right?
29:11 So let's talk about the process of maybe coming up with a new data pipeline.
29:15 What does a software project look like for you all?
29:17 Yeah.
29:18 So it usually begins with a collaborative meeting with some experimental scientists
29:24 where we discuss what's the experimental design going to be like?
29:28 What are we going to be looking at in the data?
29:30 And then the experimentalists will go and they'll generate some form of sequencing data.
29:36 At that point, we generally take the data, open up some Jupyter notebooks or
29:42 some small Python, sometimes even just bash scripts to try to use some of those
29:47 standard third-party tools.
29:49 These are things like making sure all the sequences are aligned to each other so that
29:53 you know when there's differences and making sure the quality is correct, things like
29:58 that.
29:58 These are pretty standard bioinformatics things.
30:02 Right.
30:02 Then for sequencing assays, there's usually a couple spots where there's some real
30:09 experimental logic going into it where often we'll have to write custom code in Python
30:15 to say, if there's this sequence here, it means we should keep the sequence.
30:20 And if there's this sequence here, we should divide the sequence in half or
30:23 something like that.
30:24 And so that code gets written in Python, maybe it's in the Jupyter notebook or another
30:29 script.
30:30 And we sort of do this really slow testing depending on the size of the data.
30:34 It might be locally on a laptop or in a small cloud-based HPC type cluster.
30:40 Right.
30:40 This is like where we're doing...
30:42 You're not trying to process all the results.
30:44 You just want to spot check and see if it's coming out right before you turn it loose,
30:48 right?
30:48 Right.
30:49 Or we're very patient.
30:50 It's a little bit above.
30:52 Sometimes it's very difficult to take only a small fraction of the data, but we try when we
30:57 can.
30:57 Once we settle on something that we think is pretty locked down, we'll take it out of the
31:02 Jupyter notebooks.
31:03 We don't try to use Paperville or anything like that.
31:06 We try to get it out of there as soon as possible into some more complex scripts.
31:11 They might be a shell script that runs a number of other scripts in order,
31:15 or we might start using some sort of workflow manager.
31:20 workflow managers in bioinformatics are pretty common because everyone has the same problem of
31:26 running all these third party tools together and custom code.
31:29 Right.
31:30 It's a lot of shared tools as well, probably.
31:32 Right.
31:32 They all use this library or that app.
31:34 Absolutely.
31:34 Yeah.
31:34 There's definitely...
31:36 Whatever it is, right?
31:37 There's a whole bunch of standard bioinformatics tools that we run on almost everything.
31:42 And so some of the workflow managers are designed to specifically work very well with those
31:47 tools and others are pretty agnostic of what you're doing with them.
31:52 Yeah.
31:52 One of the things I find interesting and listening to you talk about this,
31:56 it just reminded me, is so often we see these problems that people are solving, right?
32:02 Like, oh, here we're using CRISPR to do all this work.
32:06 And then you talk about the tools you use.
32:07 It's like, oh, yeah, we're using like NumPy and Pandas and Jupyter and these kinds of
32:12 things.
32:12 And the thing that I find really interesting is for software development,
32:17 there's so much of the stuff that is, it's just the same for everyone, right?
32:22 They're doing the same thing.
32:23 And then there's 10 or 20% that like, yeah, this field does this part different, but there's
32:28 like 80% of, yeah, we kind of, we should use source control.
32:32 We're using Python.
32:33 We're using notebooks.
32:34 We're using Pandas and that kind of stuff.
32:37 and it's the similarities are way more common than I think they appear from
32:43 the outside.
32:44 It's a great point.
32:45 And I think we'd, we'd all be better off if we reminded ourselves of that more often that
32:49 we're not just because we're doing biotechnology and things with Python that we're largely very
32:57 similar to other software developers doing data science on business topics or finance or
33:03 just standard web development.
33:05 You could be at a hedge fund and you're like, this is that different than what I'm used to
33:09 actually.
33:09 Yeah.
33:09 And if you think like that, we, you end up with, I think, better practices overall in
33:14 software.
33:14 Yeah.
33:15 Oh, I totally agree.
33:16 Also, I suspect that this fact is what also makes the data science tools and all the
33:23 tooling around Python, the libraries and whatnot.
33:25 So good because it's not just like, oh, the biologists have made this part of it really good.
33:30 It's like most of the people are all really refining like the core parts.
33:36 Right.
33:36 Yeah.
33:36 I think that's, that's definitely the case.
33:38 And some of the biology specific tools are, are a little, a little walkie when you, when
33:43 you start using them.
33:43 I think, I mean, things like pandas are, are really amazing.
33:47 You can tell the amount of attention or something is different.
33:50 Cool.
33:51 what is moving to production look like for you.
33:54 So you talked about sometimes it's, you start the exploration and stuff in notebooks, which is
33:58 exactly what they're built for.
33:59 And then moving to maybe a little more composition of scripts and whatnot.
34:04 And eventually somehow you end up with, you know, Lambda, cloud databases,
34:08 things like that.
34:09 What's that flow?
34:10 Yeah.
34:11 So the process of, I guess we say like productionizing a pipeline is we've had pretty well
34:17 set now.
34:17 and generally how it works as we say this pipeline is about done and we'll hand it off to
34:25 myself or one of my colleagues to start the process of getting it fully cloud capable and
34:31 scalable.
34:32 And what that means for us is to take the software in whatever form we've
34:37 gotten it from our colleagues and put it into a workflow manager.
34:42 And we, I think every company has their own version of workflow manager that they, they
34:47 choose.
34:47 we're using Luigi, which is fully Python based.
34:51 It was originally developed at Spotify to do this sort of task.
34:57 It uses like a GNU make type target file dag creation.
35:02 I don't know all the technical terms to describe how the tasks are built, but you
35:07 essentially, you have a task at the end and you say it requires the output of this other
35:13 task.
35:13 And then that task requires the more.
35:16 And as they build up, you create a graph of what tasks need to be done to get to our
35:21 output.
35:22 Right.
35:22 A directed acyclic graph.
35:24 And then the workflow can decide like, oh, these two things are independent.
35:28 So let's scale them out separately.
35:30 now this one has to wait for these two to finish and then get its results.
35:33 And that coordination can be really tricky.
35:35 Exactly.
35:36 And there's a number of common workflow managers and bioinformatics.
35:39 I think the two most common are snake make and next flow.
35:43 Luigi is, has also been really good for us.
35:47 We like it primarily because it is fully Python based and it uses standard Python syntax,
35:52 which allows us to really, if we need to, get out of the hood and add some customization,
35:58 extend it where we need to, or, or fix things that we don't like about it.
36:03 And that was a really important part of our decision and choosing Luigi over some of these
36:07 other workflow managers.
36:09 Yeah, for sure.
36:10 I had a nice conversation with the Airflow, Apache Airflow folks, and not
36:14 too long ago.
36:15 And one of the things that really struck me about this is the ability for people to work on
36:21 little parts of the processing, a little bit like that, little Python automation tools or a
36:26 little Python projects that you described earlier in episode 327 in that instead of
36:32 trying to figure out all this orchestration, you just have to figure out, well, this little
36:36 task is going to do a thing.
36:37 And that, like I said, maybe it means see the file and then copy it over there.
36:42 And if your job was see a file here, copy it over there, like that's a really simple
36:47 job.
36:47 You can totally nail that.
36:49 You know what I mean?
36:49 Yeah.
36:50 Whereas if your job is to orchestrate this, this graph and make sure that it runs on time
36:55 and here's the failure case, all of a sudden that becomes super, super hard.
36:58 So these seem really empowering, almost like the promise of microservices is like
37:03 you get to focus on one little part and do that.
37:05 Yeah, it definitely, it definitely helps.
37:08 And it helps with that idea of having these small tasks.
37:11 It really helps with how you can develop it and reuse the components for each task.
37:18 Like we might, as we said earlier, that there are these third party tools that end up being
37:21 used in almost all of our pipelines and using something like Luigi or any workflow manager.
37:27 You can reuse the tasks in different contexts as need be and you can have
37:33 your perfectly optimized way of using that task everywhere.
37:37 That reuse is really nice and it's something I think a lot of software developers appreciate.
37:41 Yeah, for sure.
37:42 So if you look at some of the folks that are using Luigi, so Spotify, as you
37:47 said, created it, but also Foursquare, Stripe, Asana, SeatGeek, a lot of companies
37:54 that people probably heard of like, oh, these places are doing awesome stuff.
37:58 Let's be like them.
37:58 Yeah.
37:59 A lot of places use it for like Hadoop and things like that.
38:03 And one of the nice things is you mentioned about like how Airflow has the same model
38:08 where you can create these contributions, which are different connectors for
38:14 Luigi or for Airflow, where you can connect them to their different cloud
38:18 providers or different data stores, things like that.
38:21 And that allows you to use Luigi or any workflow manager in numerous different
38:27 contexts, whether it's locally on your own computer running things in Docker containers or whether
38:33 it's deploying out to AWS and scaling massively horizontal.
38:38 These workflow managers really support that.
38:41 And that's why they're a necessary component in how we productionize our data
38:45 pipelines.
38:46 Yeah.
38:46 Again, they just seem so empowering for allowing people to focus on just
38:50 each step independently, which is excellent.
38:53 Did you consider other ones like did you consider Airflow or DAXer or any
38:57 of these other ones or did you find this fit and you're like, we're going
39:00 with this?
39:01 We did look at some other ones.
39:03 We were using Nextflow for a little bit, which is a bioinformatic flavored workflow
39:10 manager.
39:10 It's very focused on bioinformatics as its primary use case, although you could use it
39:15 for anything.
39:16 Syntax is similar to Groovy and it's based in Groovy.
39:20 And that was one of the detractives for us is that it was a little hard to get
39:24 under the hood and use that because of it.
39:27 I did briefly look at DAXer after hearing a few episodes, I think.
39:31 No, maybe a different podcast.
39:32 Yeah, I did have Tobias Macy on to give us an overview of the whole data
39:36 engineering landscape.
39:37 So possibly, I know he spoke about it then, but I'm not sure when you heard
39:41 about it.
39:41 Yeah, so I heard about it on a podcast and probably this one as well.
39:44 And I did look into it, but it didn't have it at that time.
39:48 It was pretty early and it didn't have any connectors to AWS in the ways that we
39:53 like to use.
39:54 The connectors, right?
39:55 That's such an important thing because otherwise you've got to learn the API
39:59 of every single thing you're talking to.
40:01 Yeah.
40:01 These days, knowing how Luigi works, it actually wouldn't have been that big
40:05 of a task to look under the hood.
40:07 But yeah, so we did choose Luigi and particularly we like how it handles
40:13 deployment to AWS and we use it on the service called AWS Batch, which is, I guess it might be
40:22 similar to like Kubernetes pod, although I haven't done anything with K8s or anything like that.
40:29 so I'm not speaking from experience, but it essentially scales up EC2 instances, these elastic
40:36 compute instances on the cloud as you need them.
40:41 And it gives out jobs to the virtual computers as necessary.
40:46 So it spins them up, allocates jobs in a Docker container.
40:50 They run when there's no more jobs on the instance, it shuts off.
40:54 Okay.
40:55 So you come up with like an AMI, an Amazon machine image that's pre-configured,
41:00 set up, ready to run.
41:01 And then you say, I'm going to give you a bunch of data.
41:04 Each one of these data gets passed to the machine and it runs and then maybe even
41:08 shuts down when it's done.
41:09 Yeah.
41:10 There is an AMI.
41:11 We keep the AMI pretty simple because it's the sort of the base for all of
41:15 them.
41:15 And then the way batch works is you have your top level AMI that's called the
41:20 compute environment, I believe.
41:22 And then inside of it, you run the actual job and the job runs inside of a
41:28 Docker container.
41:29 Oh, I see.
41:29 So the Docker container is pre-configured with like all the Python dependencies and
41:34 the settings that it needs and whatnot.
41:35 Right.
41:36 So we have each task in Luigi, each little piece of work as its own Docker
41:43 container.
41:43 and then we push those out into the cloud and they get allocated out onto these
41:49 machines.
41:50 They run their task.
41:51 Data comes in from S3, goes back out to S3.
41:55 Nothing is left on the hard drives and then they disappear.
41:59 They're these little ephemeral compute instances.
42:02 And that's all managed by a workflow manager such as Luigi or Airflow or
42:07 Nexflow.
42:07 Wow.
42:08 That's pretty awesome.
42:09 One of the things that I remember reading and thinking that's a pretty crazy use of the cloud was
42:16 this Ars Technica article from look at that year, 2011.
42:21 Oh, wow.
42:21 So if you think back to 2011, like the cloud was really new and the idea of
42:27 spending a ton of money on it and getting a bunch of compute out of it was still
42:31 somewhat foreign to people.
42:32 So there's this article I'll link to called the 100 or 1,279 per dollar per hour,
42:39 30,000 core cluster built on Amazon EC2 cloud, which is this company, this
42:46 pharmaceutical company that needed to do a lot of computing.
42:49 And they said, instead of buying a supercomputer, basically, we're going to
42:54 come up and just fire off an insane amount of cores.
42:57 And I think if I remember reading this correctly, that they weren't allowed to make
43:01 that many cores in a single data center.
43:05 so they had to do this across.
43:07 They also had to coordinate in like multi data center type of processing as well
43:12 because, yeah, it's just the scale of everything.
43:15 This seems like the type of work that you all might be doing.
43:18 Yeah, it does look very familiar.
43:21 Okay.
43:22 You got any war stories?
43:24 Can you tell us anything about this?
43:25 Yeah, we do occasionally have a certain type of molecular modeling job that we can scale
43:31 very wide.
43:33 And I think these, this sort of 30,000 number looks pretty familiar.
43:38 I think our largest jobs today have been about 10,000 CPUs wide and running for a
43:46 few days.
43:47 So maybe like four or five days.
43:48 I think the number was like four or five days on the 10,000 cores.
43:52 Yeah.
43:53 Yeah.
43:53 It's a, it's, it's a lot.
43:56 And I, so I think it was like over a million CPU hours on AWS batch.
44:01 And that was just something that we could really heavily parallelize and we needed the
44:08 data fast and it worked.
44:10 You pull it all back and aggregate it all together at the end that it was a really useful
44:16 data set.
44:17 And it's pretty amazing what you can do on some of these cloud providers by going really wide.
44:23 Yeah.
44:23 It's crazy.
44:25 You know, I mean, there's some places where it makes sense to like build true supercomputers
44:29 like Oak Ridge has this thing called summit, which is like this insane supercomputer that
44:35 they have.
44:35 But a lot of times there's the latency of getting something like this set up.
44:40 There's the overhead of, guess what?
44:42 Now you're an admin for like an insane, there's only three in the world supercomputer type
44:47 system.
44:47 It's got to be empowering to be able to just go hit go and then let all this happen and not worry
44:53 about all those details.
44:54 Right.
44:55 Yeah.
44:55 And there's definitely still some stress involved in, in starting one of these jobs.
44:59 If you just, you know, it's not cheap anyway, slice it.
45:03 We do, we do try to, you know, do everything we can to make it as, as efficient in cost as
45:08 possible.
45:08 But yeah, I'm sure there's, there's two aspects to it that really jump out to me.
45:12 One is the, Oh, was that the wrong version of the code?
45:15 The one that still had the bug?
45:17 We got to throw away all the data.
45:18 Whoops.
45:18 We just ran for five days and, you know, thousands of dollars got burned up and there's a bit
45:24 of a problem there.
45:25 So whoops.
45:26 And now it's five days later until you get the answer as well.
45:29 Right.
45:29 That's one.
45:31 And then I'm losing track of my thought on my other one, but yeah, anyway, it's just got to be
45:36 stressful to be like, to set that up and then press go.
45:39 Right.
45:40 it definitely is.
45:41 I think one of the benefits of a lot of our computational team does have a spare metal background and
45:46 doing experiments in the lab, these sort of numbers, like tens of thousands of dollars for an
45:52 experiment are not really that uncommon.
45:54 And those kinds of things, you get really careful and you check all the boxes and double check
45:59 everything.
45:59 And so I think a lot of us have had that, that experience.
46:02 And so even when we're dealing with software, we'll be very careful and we'll do the tests and
46:08 quality control before we really let her rip.
46:10 Yeah.
46:10 I, you know, I worked on some projects when I was in grad school that were on a Silicon graphics, big
46:16 mainframe type thing.
46:17 And obviously much lower importance than, you know, solving diseases and
46:23 stuff is just solving math problems.
46:24 But I remember coming in to work on the project one day and none of our workstations could log in to the
46:31 Silicon graphics machine.
46:33 And what is wrong with this thing?
46:34 And it was so loud.
46:35 It was in the other room.
46:36 You could hear it roaring away.
46:38 in there.
46:38 It clearly was loud.
46:40 And what happened was it wasn't me.
46:42 It was someone else in the group had written some code.
46:47 And these, these things would run all night.
46:48 We'd come in in the morning and we'd check them.
46:49 And what had happened was they had a bug in their code, which they knew they were trying to diagnose
46:55 it.
46:55 So they were printing out a bunch of log stuff or something like that.
46:58 Well, they did that in a tight loop on a high-end computer for a night.
47:03 And it filled up the hard drive till it had zero bytes left.
47:06 And apparently the Silicon graphics machine couldn't operate anymore was literally zero bytes.
47:12 And so it just stopped working.
47:13 They couldn't get it eternal.
47:14 I was like, it took days to get it back, I believe.
47:16 Oh, oops.
47:18 But it's like that kind of stuff, right?
47:20 I mean, this, you're not going to break EC2, but, you know, you don't know until the next day that, oh, look,
47:25 you filled up the computer and it doesn't work anymore, right?
47:27 When you're doing that much computing, you could run out of different resources.
47:31 You could run into all kinds of problems.
47:33 Absolutely.
47:34 And we are without our worst stories of doing this, but I think we've definitely learned a lot of lessons
47:41 along the way of how to monitor your jobs effectively and double check things.
47:46 But sometimes, yeah, you run a big job and it doesn't quite turn out right.
47:51 But it's the cost of doing business, I guess.
47:53 Yeah.
47:53 Yeah.
47:54 I mean, it's very computational to explore stuff like this kind of stuff.
47:57 But that's also what enables it, right?
47:59 Without this computing power, it would just not be a thing.
48:02 Absolutely.
48:02 A lot of the data just takes a lot of time to process and there's just not
48:06 really any way around it.
48:07 And even when you're iterating, you have to go through all the hoops to look at the data at the end.
48:12 Yeah.
48:12 So you talked about APIs.
48:14 You talked about data store.
48:16 What are you using for a database?
48:18 Is this like a hosted RDS AWS thing or what is the story with that?
48:24 Yeah.
48:24 So we have a few different places to store data.
48:27 our larger scale internal data, we store in Django-based web app and we use the Django ORM for SQL-based
48:37 database, so MySQL database on AWS.
48:40 And that has worked surprisingly effectively, actually.
48:44 I've heard some people say that the Django ORM is really slow when you scale out and things.
48:50 But if you design it correctly, I think it'll surprise you.
48:55 I think that's so true.
48:57 I hear so many things about ORMs are slow or this thing is slow in this way.
49:02 And wow, if you have the queries structured well, you do the joins ahead
49:07 of time.
49:07 If you have indexes and you've put the work into finding all these things, it's
49:12 mind-blowing.
49:12 When I go to sites, I won't call any out.
49:15 I don't know if they've been updated or whatever.
49:17 But you go to a site and you're like, this site is taking four or five seconds to load.
49:20 What could it possibly be doing?
49:22 I mean, I know it has some data, but it doesn't have unimaginable amounts of data.
49:27 Right?
49:27 Surely somebody could just put an index in here or a worst case, a cache in something.
49:32 And it would just transform it.
49:34 Right?
49:34 So yeah, I'm glad to hear you're having good experiences.
49:37 Yeah, we definitely fairly regularly run into slow queries.
49:42 They're usually not too bad to solve.
49:44 I'm sure at some point we'll get to something really wonky that will be challenging.
49:48 But I think for the most part, yeah, we've been able to solve it through better query design
49:54 and better indexing.
49:56 Yeah.
49:56 Do you ever do things where you opt out of the sort of class-based query syntax and go
50:02 straight to SQL queries here and there to make sure that that part works better?
50:07 We have tried it for some particular sequence-based searches that we do.
50:13 And I actually found that most of the time you can write it in the ORM.
50:17 It's just a little more complicated.
50:19 But I do expect that at some point we will be writing raw SQL queries because out of necessity.
50:25 Yeah, but it's not the majority.
50:26 Mainly you're using the ORM and then it's okay.
50:29 Yeah.
50:29 And I think a lot of benefits from the appropriate data model will help the queries along the way.
50:35 Yeah, absolutely.
50:36 The thing I found the slowest about ORMs and ODM, if you're doing document databases,
50:41 is the deserialization, actually.
50:44 It's not the query time, but it's like, I got to generate 100,000 Python objects
50:49 and that just happens to be not that fast relative to other things.
50:53 You're talking about my Monday morning pipeline.
50:58 And in those cases, I think that's the place where it makes sense to maybe do some kind of like a projection or something.
51:05 I don't know how to do it in Django ORM, but in say Mongo Engine, you can say,
51:09 I know what I'm going to get back is an iterable set of these objects that match to the data,
51:15 but only actually add, just fill out these two fields.
51:18 Most of the data, just throw it away.
51:20 Don't try to parse it and convert it.
51:22 Just like, I just want these two fields.
51:24 And that usually makes it dramatically faster.
51:26 Yeah, we've run into a number of bottlenecks at the serialization layer.
51:31 And we have been experimenting with a variety of different ways to solve those issues.
51:37 And sometimes it means putting fewer Python objects between you and the data.
51:42 And that often speeds it up, even if it makes it a little bit harder to interpret in your development environment.
51:49 Yeah, absolutely.
51:50 Or just say, you know what?
51:51 I just need dictionaries this time.
51:53 I know it's going to be less fun, but that's what it takes.
51:56 That was the fix on Monday morning.
51:58 We do try to extensively use data classes for a lot of our interoperability.
52:04 When data comes in and out of a pipeline, we like to have it in a data class, in a centrally stored repository.
52:11 And then our Django web app also has access to that.
52:15 So it knows what the structure of the data coming in is, and it knows what to serialize it to when it's coming out.
52:20 And the Python data classes has been a really useful tool for that.
52:25 But I think we were talking about that on another podcast a few weeks ago.
52:30 Maybe it was the Python bytes one.
52:31 The data classes can be slow.
52:33 And sometimes it's better to just have a dictionary, even if it is a very highly structured dictionary.
52:40 Yeah, and the problem is maintainability and whatnot.
52:43 But, you know, if it's five times slower, you know what?
52:45 This time it matters.
52:47 So we're going to just bite the bullet and have to deal with it.
52:49 Let's see.
52:50 We had time for just a little bit more digging into it.
52:52 So you talked about the Django ORM, Django REST framework, which is all great.
52:56 What's the server deployment story?
53:00 Like, how do you run that thing?
53:01 Is it with G-Unicorn?
53:03 Is it Micro Whiskey?
53:04 You know, what's your setup on that side of things?
53:07 Ours is a little custom.
53:10 I guess in some ways it's pretty standard.
53:12 I think we're blanking on exactly how it's set up now.
53:16 But there's an NGINX proxy I am blanking on.
53:21 It might be G-Unicorn.
53:22 I feel like G-Unicorn and Django go together frequently.
53:25 I'm not sure why they got paired up specifically.
53:28 But yeah, it's a good one.
53:30 And we end up deploying it out to AWS Elastic Beanstalk, which is a source of some conversation in our team.
53:39 Because there's some things we really like about it.
53:41 And there's some things that are really annoying in terms of the deployment is much more complicated than we would like it to be.
53:47 But we have everything wrapped up in a pretty gnarly CDK stack that does a lot of this work.
53:54 It was messy, but you've solved it with a CDK.
53:57 Now it's just you push the button and it's okay.
53:59 It's exactly like that.
54:00 We have a very automated deployment process.
54:03 I wouldn't like to refactor it, but it's there and it works.
54:09 So that works for us.
54:10 But I think, yeah, it's a pretty standard Django deployment on the cloud.
54:14 And it works well.
54:16 Yeah, cool.
54:17 All right.
54:17 Well, I think that's probably about it for time to talk about the specifics of what you all are doing.
54:22 But there's the last two questions, as always.
54:25 So let's start with the editor.
54:27 If you're going to write some Python code, what editor do you use?
54:30 The yes code.
54:31 A lot of the remote development environment on that.
54:34 Oh, yeah.
54:34 You use the remote aspect of it?
54:37 Yeah.
54:37 We're doing a lot of work on EC2 instances as our day-to-day work.
54:41 And the yes code, the way it works with instances in the cloud is really amazing.
54:47 So I would encourage anyone to check out that extension.
54:51 Yeah, you get access to the file system on the remote machine.
54:55 And basically, it's just your view into that server.
54:58 But it's more or less standard VS Code, right?
55:00 But when you hit run, it just runs up there.
55:02 It feels exactly like you're on your own computer.
55:04 Sometimes I actually get confused whether I'm on or not.
55:07 Oh, so does it work?
55:08 Because I'm in Virginia.
55:09 I see.
55:10 Yeah.
55:10 All right.
55:10 And then notable PyPI package?
55:12 I'll have to shout out some of the ones that we talked about.
55:14 I would encourage people to look at AWS CDK.
55:17 If they're on AWS, I think it has some really interesting things there.
55:21 And then also, Luigi as a workflow manager, if people are doing any of these types of data pipelines
55:28 that have tasks that get reused, these sort of workflow managers are really cool.
55:31 And Luigi is a pretty accessible one, probably one that's familiar with Python.
55:35 Yeah.
55:35 All right.
55:36 Fantastic.
55:36 Yeah.
55:37 I'm just learning to embrace these workflow managers, but they do seem really amazing.
55:41 All right.
55:42 So for all the biologists and scientists out there listening, thinking you've got this really
55:47 cool setup and all this cool computational infrastructure, what do you tell them?
55:50 How do they get started?
55:51 Maybe biology or wherever?
55:54 I think biology is a good place to start.
55:55 We're also happy to have people come from a software background that are really interested
55:59 in learning the biology.
56:00 And I guess as a final plug, we do have a few open positions.
56:04 So if you're interested, check out our careers page and give us an application.
56:09 Are you guys a remote place?
56:11 Remote friendly?
56:13 Or what's the story these days?
56:14 Yep.
56:14 We're remote friendly.
56:15 I'm actually living in Philadelphia and our company is based in Cambridge.
56:19 So we are.
56:20 I mean, it's a short trip to the cloud, no matter where you come from, basically.
56:25 Absolutely.
56:25 Right on.
56:26 David, thank you for being here and giving us this look inside all the gene editing Python
56:32 stuff you're doing.
56:33 Thank you, Michael.
56:33 It was a pleasure.
56:34 Yeah, you bet.
56:34 Bye.
56:35 Bye.
56:35 This has been another episode of Talk Python To Me.
56:39 Our guest on this episode was David Bourne, and it's been brought to you by Shortcut, us
56:44 over at Talk Python Training, and the transcripts were brought to you by Assembly AI.
56:48 Choose Shortcut, formerly Clubhouse.io, for tracking all of your project's work, because you shouldn't
56:54 have to project manage your project management.
56:57 Visit talkpython.fm/shortcut.
56:59 Do you need a great automatic speech-to-text API?
57:03 Get human-level accuracy in just a few lines of code.
57:05 Visit talkpython.fm/assembly AI.
57:09 Want to level up your Python?
57:10 Want to level up your Python?
57:10 We have one of the largest catalogs of Python video courses over at Talk Python.
57:14 Our content ranges from true beginners to deeply advanced topics like memory and async.
57:19 And best of all, there's not a subscription in sight.
57:22 Check it out for yourself at training.talkpython.fm.
57:25 Be sure to subscribe to the show.
57:27 Open your favorite podcast app and search for Python.
57:29 We should be right at the top.
57:31 You can also find the iTunes feed at /itunes, the Google Play feed at /play, and the direct
57:37 RSS feed at /rss on talkpython.fm.
57:41 We're live streaming most of our recordings these days.
57:44 If you want to be part of the show and have your comments featured on the air, be sure to subscribe to our YouTube channel at talkpython.fm/youtube.
57:51 This is your host, Michael Kennedy.
57:53 Thanks so much for listening.
57:55 I really appreciate it.
57:56 Now get out there and write some Python code.
57:58 I really appreciate it.